The BnF Gallica image library contains thousands of freely accessible images of historical interests. It provides an Application Programming Interface to access it (see their documentation). The access is possible through the IIIF API. The goal of bnfimage is to let you access this API directly from R.
Get Image(s) - bi_image()
To get a specific image, you first need to get its identifier from Gallica. The identifier is a string that begins with ark:/. It is accessible from browsing Gallica as the URL of the object such as: https://gallica.bnf.fr/ark:/12148/bpt6k10411495/f11.item# giving the id ark:/12148/bpt6k10411495/f11.
Single Image
To download a single image you can use the bi_image() with the identifier of the object:
bi_image("ark:/12148/bpt6k10411495/f11")
plot of chunk bi-image
Image Crop
This displays a small portion of the image as by default this functions only returns the top left corner of the image from 0 to 500px in both directions.
To get the full image you can use the argument region with the value "full":

plot of chunk bi-image-region-full
Otherwise if you want to crop a specific region in the image you can define it as length 4 numeric vector specifiying the coordinates of the top left most point and the size of the extracted region in the form x0, y0, w, h. For example if you want to extract the image from 1250px x and 1300px y of 60px large and 50px high you would call:

plot of chunk bi-image-region-crop
Image size
As you noticed previous commands specified a size argument. It corresponds to the size of the extracted image. It can be equal to "full" so that the extracted region is returned without being scaled.

plot of chunk bi-image-size-full
It can also be a numeric vector of lenth 2 that gives the size at which the extracted region should be scaled:

plot of chunk bi-image-size-small
Other options exist in the IIIF Image API but are not yet implemented in bnfimage.
Rotation
bnfimage supports a rotation argument that returns a rotated image. By default the returned image is not rotated (rotation = 0). The argument is the clockwise rotation angle in degrees. When the rotation angle is not a multiple of 90° then the background color used is white.
# No rotation
bi_image("ark:/12148/bpt6k10411495/f11", region = c(1250, 1300, 1600, 1650),
size = c(40, 41),
rotation = 0)
plot of chunk bi-image-region-crop-rotation
# 90° clockwise rotation
bi_image("ark:/12148/bpt6k10411495/f11", region = c(1250, 1300, 1600, 1650),
size = c(40, 41),
rotation = 90)
plot of chunk bi-image-region-crop-rotation
# Non-multiple of 90° rotation
bi_image("ark:/12148/bpt6k10411495/f11", region = c(1250, 1300, 1600, 1650),
size = c(40, 41),
rotation = 153)
plot of chunk bi-image-region-crop-rotation
Color
To lower bandwidth it is possible to directly ask for transformed bit-depth of the returned image through the quality argument. It can take four values:
-
"native"which returns the image as stored in the BnF database, -
"color"with the image in full color using 24 bits per pixel, -
"grey"with the image in greyscale using 8 bits per pixel, -
"bitonal"where is pixel is either black or white using 1 bit per pixel.
# Native
bi_image("ark:/12148/bpt6k10411495/f11", region = c(1250, 1300, 1600, 1650),
size = c(48, 50),
quality = "native")
plot of chunk bi-image-quality
# Color
bi_image("ark:/12148/bpt6k10411495/f11", region = c(1250, 1300, 1600, 1650),
size = c(48, 50),
quality = "color")
plot of chunk bi-image-quality
# Grey
bi_image("ark:/12148/bpt6k10411495/f11", region = c(1250, 1300, 1600, 1650),
size = c(48, 50),
quality = "gray")
plot of chunk bi-image-quality
# Bitonal
bi_image("ark:/12148/bpt6k10411495/f11", region = c(1250, 1300, 1600, 1650),
size = c(48, 50),
quality = "bitonal")
plot of chunk bi-image-quality
Format
The images can be returned as different files format. This can be changed through the format argument which can take all these values:
-
"jpg"JPEG format (which is used by default), -
"gif"GIF format, -
"png"PNG, -
"jp2"JP2, -
"pdf"PDF, -
"tif"TIFF.
# JPG image
bi_image("ark:/12148/bpt6k10411495/f11", region = "full", size = c(75, 75),
format = "jpg")
plot of chunk bi-image-format
# PDF image
bi_image("ark:/12148/bpt6k10411495/f11", region = "full", size = c(75, 75),
format = "pdf")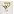
plot of chunk bi-image-format
Saving the image
To save the images you can use the function image_write() in the magick package. image_write() can accept any image filetype:
img = bi_image("ark:/12148/bpt6k10411495/f11", size = c(48, 50),
region = c(1250, 1300, 1600, 1650))
magick::image_write(img, "./my_image.png")The second argument specifies the file path. Refer to the help of the function through ?magick::image_write to get an overview of all the arguments and uses.
Multiple Images
To access several images you can use a list of identifier and wrap calls to bi_image() using lapply():
several_ids = c("ark:/12148/bpt6k10411495/f11", "ark:/12148/bpt6k10411495/f13",
"ark:/12148/bpt6k10411495/f15")
several_imgs = lapply(several_ids, bi_image, region = "full",
size = c(50, 50))
several_imgs
#> [[1]]
#> # A tibble: 1 x 7
#> format width height colorspace matte filesize density
#> <chr> <int> <int> <chr> <lgl> <int> <chr>
#> 1 JPEG 50 50 sRGB FALSE 1480 400x400
#>
#> [[2]]
#> # A tibble: 1 x 7
#> format width height colorspace matte filesize density
#> <chr> <int> <int> <chr> <lgl> <int> <chr>
#> 1 JPEG 50 50 sRGB FALSE 1154 400x400
#>
#> [[3]]
#> # A tibble: 1 x 7
#> format width height colorspace matte filesize density
#> <chr> <int> <int> <chr> <lgl> <int> <chr>
#> 1 JPEG 50 50 sRGB FALSE 1475 400x400The arguments can be specified similarly to single calls to bi_image().
Get Image(s) Metadata - bi_metadata()
One interest of the BnF image library is the richness of the metadata associated with an image. For the moment bnfimage only converts metadata to a giant list to make it accessible.
Single Image
Using a single image identifier you can retrieve the metadata of an image through the function bi_metadata(). The first argument is the identifier of the image as for bi_image():
met = bi_metadata("ark:/12148/bpt6k10411495/f11")
str(met, max.level = 1)
#> List of 13
#> $ @id : chr "https://gallica.bnf.fr/iiif/ark:/12148/bpt6k10411495/manifest.json"
#> $ label : chr "BnF, département Arsenal, GR FOL-30 (2)"
#> $ attribution: chr "Bibliothèque nationale de France"
#> $ license : chr "https://gallica.bnf.fr/html/und/conditions-dutilisation-des-contenus-de-gallica"
#> $ logo : chr "https://gallica.bnf.fr/mbImage/logos/logo-bnf.png"
#> $ related : chr "https://gallica.bnf.fr/ark:/12148/bpt6k10411495"
#> $ seeAlso :List of 1
#> $ description: chr "Les liliacées. Tome 2 / , par P.-J. Redouté. [Tome premier-] huitième"
#> $ metadata :List of 13
#> $ sequences :List of 1
#> $ thumbnail :List of 1
#> $ @type : chr "sc:Manifest"
#> $ @context : chr "http://iiif.io/api/presentation/2/context.json"The metadata contains much information on the actual image as well as its context.
Multiple Images
Similarly as for bi_image() to access the metadata of several images you can wrap the calls to bi_metadata() through a list of identifiers using lapply():
several_met = lapply(several_ids, bi_metadata)
str(several_met, max.level = 2)
#> List of 3
#> $ :List of 13
#> ..$ @id : chr "https://gallica.bnf.fr/iiif/ark:/12148/bpt6k10411495/manifest.json"
#> ..$ label : chr "BnF, département Arsenal, GR FOL-30 (2)"
#> ..$ attribution: chr "Bibliothèque nationale de France"
#> ..$ license : chr "https://gallica.bnf.fr/html/und/conditions-dutilisation-des-contenus-de-gallica"
#> ..$ logo : chr "https://gallica.bnf.fr/mbImage/logos/logo-bnf.png"
#> ..$ related : chr "https://gallica.bnf.fr/ark:/12148/bpt6k10411495"
#> ..$ seeAlso :List of 1
#> ..$ description: chr "Les liliacées. Tome 2 / , par P.-J. Redouté. [Tome premier-] huitième"
#> ..$ metadata :List of 13
#> ..$ sequences :List of 1
#> ..$ thumbnail :List of 1
#> ..$ @type : chr "sc:Manifest"
#> ..$ @context : chr "http://iiif.io/api/presentation/2/context.json"
#> $ :List of 13
#> ..$ @id : chr "https://gallica.bnf.fr/iiif/ark:/12148/bpt6k10411495/manifest.json"
#> ..$ label : chr "BnF, département Arsenal, GR FOL-30 (2)"
#> ..$ attribution: chr "Bibliothèque nationale de France"
#> ..$ license : chr "https://gallica.bnf.fr/html/und/conditions-dutilisation-des-contenus-de-gallica"
#> ..$ logo : chr "https://gallica.bnf.fr/mbImage/logos/logo-bnf.png"
#> ..$ related : chr "https://gallica.bnf.fr/ark:/12148/bpt6k10411495"
#> ..$ seeAlso :List of 1
#> ..$ description: chr "Les liliacées. Tome 2 / , par P.-J. Redouté. [Tome premier-] huitième"
#> ..$ metadata :List of 13
#> ..$ sequences :List of 1
#> ..$ thumbnail :List of 1
#> ..$ @type : chr "sc:Manifest"
#> ..$ @context : chr "http://iiif.io/api/presentation/2/context.json"
#> $ :List of 13
#> ..$ @id : chr "https://gallica.bnf.fr/iiif/ark:/12148/bpt6k10411495/manifest.json"
#> ..$ label : chr "BnF, département Arsenal, GR FOL-30 (2)"
#> ..$ attribution: chr "Bibliothèque nationale de France"
#> ..$ license : chr "https://gallica.bnf.fr/html/und/conditions-dutilisation-des-contenus-de-gallica"
#> ..$ logo : chr "https://gallica.bnf.fr/mbImage/logos/logo-bnf.png"
#> ..$ related : chr "https://gallica.bnf.fr/ark:/12148/bpt6k10411495"
#> ..$ seeAlso :List of 1
#> ..$ description: chr "Les liliacées. Tome 2 / , par P.-J. Redouté. [Tome premier-] huitième"
#> ..$ metadata :List of 13
#> ..$ sequences :List of 1
#> ..$ thumbnail :List of 1
#> ..$ @type : chr "sc:Manifest"
#> ..$ @context : chr "http://iiif.io/api/presentation/2/context.json"
Get Both Images and Metadata - bi_all_data()
The bi_all_data() function can be used to quickly retrieve both the images as well as the associated metadata of one or several images. The outputs stores the identifier of an image in a column, the actual image in a second one, and the associated metadata in a third one:
bi_all_data(c("ark:/12148/bpt6k10411495/f11", "ark:/12148/bpt6k10411495/f12"),
size = c(50, 50))
#> Error in curl::curl_fetch_memory(url, handle = handle): Timeout was reached: [gallica.bnf.fr] Connection timed out after 10002 millisecondsRate Limitation
By default bnfimage implements rate limitation through the ratelimitr package. The query are limited to a maximum of 1 every 3 seconds to avoid being blocked by the BnF server. This is the limit at which the BnF server considers the queries to be malicious. For the moment this limitation cannot be lifted.